Short Bio
The Past
I was raised in Gradisca d’Isonzo, a small town near Trieste, where I was born in 1993. After High School, in 2012, I’ve worked for a year and then I decided to move in Bologna where I got my Bachelor of Science in 2017.
In march 2020 I got my Master degree in “Biodiversity and Evolution”,
cum laude, with a dissertation entitled: Molecular
Phylogenetics, Divergence time and Evolution of Phasmida
(Insecta), supervised by professor Andrea
Luchetti. The thesis was the result of one year of
intership at the MoZoo
Lab, where I was involved phylogentic and macroevolutionary
analyses, focused on the evolution of wings in Phasmatodea (Insecta) (more info).
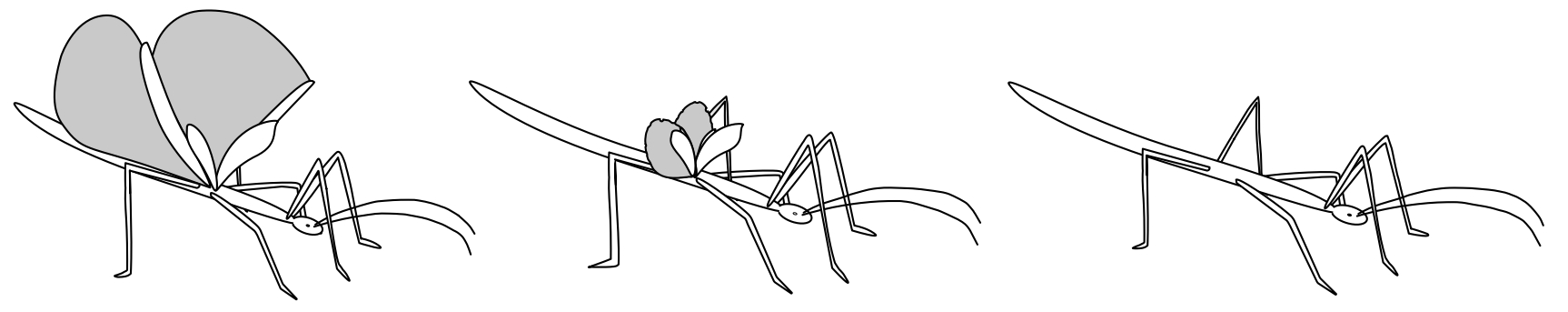
The present and the near future
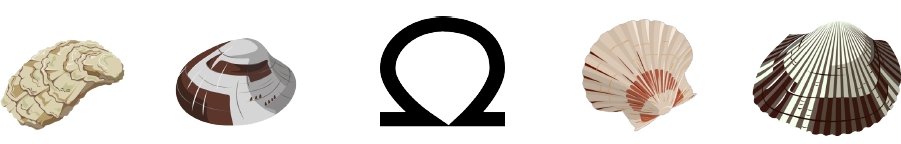
Starting from November 2020 I am a Ph.D candidate working on bivalves genomics and evolution in a comparative genomic approach, supervised by professor Fabrizio Ghiselli at the MoZoo Lab. My main focus are Transposable elements (TEs) characterization and their evolutionary dynamics trying to understand how these elements have shaped bivalve genome evolution.
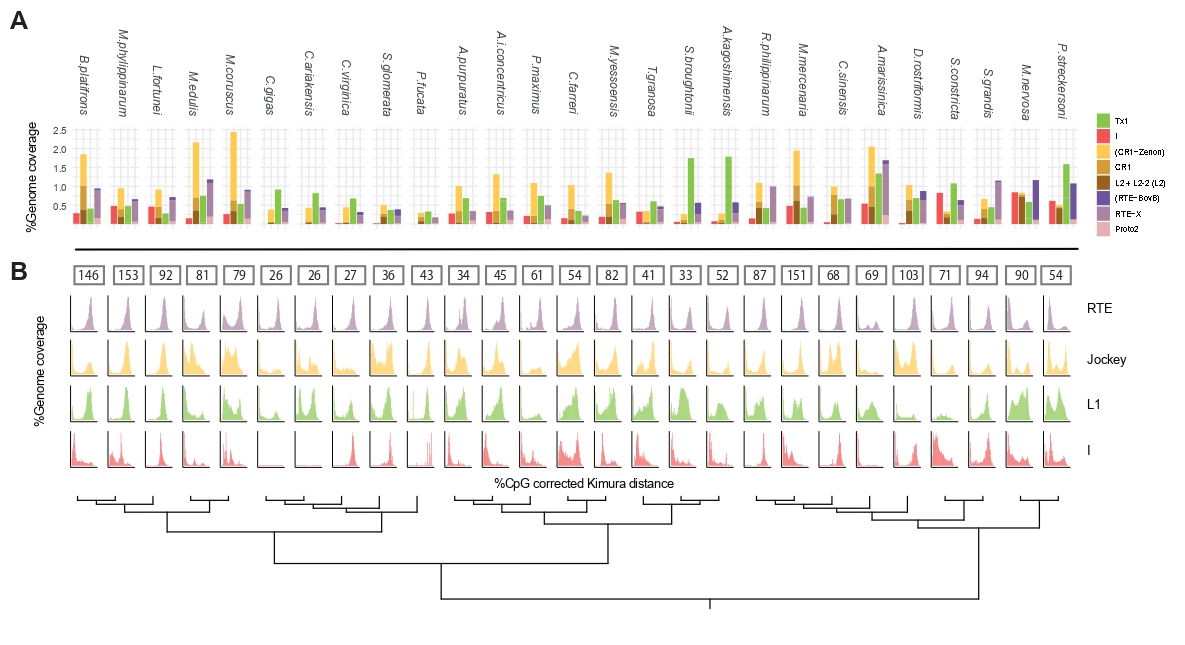
Moreover, I am involved in comparative genomics studies of Notostraca and in the termite Reticulitermes lucifugus.
Short CV
Courses:
- Comparative genomics; Online (21-25/09/20); Physalia course.
- First Step with Python in Life Science; Online (22-24/03/21); Swiss
Institute of Bioinformatics.
- Analysis of transposable elements; Online (21-25/06/21); Physalia course.
Conferences:
-Jacopo Martelossi, Filippo Nicolini, Fabrizio
Ghiselli, Andrea Luchetti. Multiple LINE lineages contribute to early
and recent bivalve genome evolution. Poster Presentation. European
Society for Evolutionary Biology (ESEB). 08-2022.
-Jacopo Martelossi, Filippo Nicolini, Fabrizio
Ghiselli, Andrea Luchetti. Multiple LINE lineages contribute to early
and recent bivalve genome evolution. Oral presentation. Italian
Society for Evolutionary Biology (SIBE). 09-2022.
-Jacopo Martelossi, Filippo Nicolini, Fabrizio
Ghiselli, Andrea Luchetti. Multiple LINE lineages contribute to early
and recent bivalve genome evolution. Poster Presentation.
Biodiversity Genomics conference. 10-2022.
Publications:
-Forni, G., Cussigh, A., Brock, P.D., Jones, B.R., Nicolini, F.,
Martelossi, J., Luchetti, A., Mantovani, B., 2022.
Taxonomic revision of the Australian stick insect genus Candovia
(Phasmida: Necrosciinae): insight from molecular systematics and
species-delimitation approaches. Zoological Journal of the Linnean
Society zlac074. https://doi.org/10.1093/zoolinnean/zlac074.
-Forni, G.1, Martelossi, J.1,
Valero, P., Hennemann, F.H., Conle, O., Luchetti, A., Mantovani, B.,
2022. Macroevolutionary Analyses Provide New Evidence of Phasmid Wings
Evolution as a Reversible Process. Systematic Biology syac038. https://doi.org/10.1093/sysbio/syac038.1
Equal contribution.
-Luchetti, A., Forni, G., Martelossi, J., Savojardo,
C., Martelli, P.L., Casadio, R., Skaist, A.M., Wheelan, S.J., Mantovani,
B., 2021. Comparative genomics of tadpole shrimps (Crustacea,
Branchiopoda, Notostraca): Dynamic genome evolution against the backdrop
of morphological stasis. Genomics 113, 4163–4172. https://doi.org/10.1016/j.ygeno.2021.11.001.
Prizes
-SIBE Award 2022 for the article: “Macroevolutionary Analyses Provide New Evidence of Phasmid Wings Evolution as a Reversible Process” (Systematic Biology 2022; syac038)